Microbial Physiology
Studying physiology and metabolism of microbes.
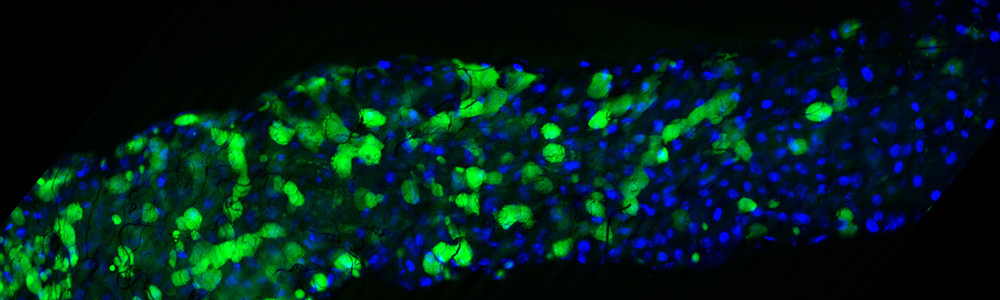
Composition and development of the characteristic Thermotoga maritima cell sheath or "toga"
Dr. Noll’s laboratory studies the physiology and molecular genetics of hyperthermophilic bacteria and archaea to shed light on their mechanisms of evolution, cellular structure, and potentials for biofuel generation. Strictly anaerobic, extremely thermophilic bacteria belonging to the group Thermotogales are the primary organisms under study. Our evolutionary studies focus on how the evolution of sugar transporters including their horizontal transfer among bacteria and archaea affects the binding properties of their proteins. Another research focus involves the unique cell envelope found in the Thermotogales, a structure with features perhaps reminiscent of cell barriers found in ancestors of bacteria and archaea. Finally a new effort to engineer thermophiles for use in biofuels generation is underway, an effort that seeks to address some of the major impediments to biomass utilization. Full genome sequences for several members of the Thermotogales have become available due to the efforts of our lab and this information is being used to guide these studies. Genomic and proteomic approaches are being used for these projects.
Genomics, proteomics and transcriptomics of bacteria (actinomycetes) that infect higher plants and fix atmospheric nitrogen; diversity of fungi associated with surface ripened cheese
Benson's research interests include microbial biogeography, including the molecular and genomic interactions of bacteria with their environment, and the genomic evolution controlling distribution of microbes in several environments. The specific areas of interest include plant-microbe and insect-microbe microbiomes, and the distribution and diversity of microorganisms associated with cheese.
Mechanisms and Regulation of Bacterial Spore Germination
Dr. Peter Setlow’s laboratory is working on the mechanisms of the germination of spores of Bacillus and Clostridium species, including the structural biology of proteins involved, interactions between germination proteins, factors regulating rates of spore germination, and the signal transduction pathways important in the overall germination process.
Physiology of methane-producing archaea
Methane-producing archaea (also known as methanogenic archaea or methanogens) generate about a billion tons of methane each year in free-oxygen environments (sediments, semi-aquatic environments and intestinal tract of animals). Methanogens are crucial in nutrient cycling and balance the Earth's climate because methane is the final product in the breakdown of organic matter, but also a potent greenhouse gas. The Microbial Ecophysiology Lab (Santiago-Martinez Lab) focuses on understanding the regulation of metabolic pathways and other cellular processes in methane-producing archaea and how energy status influences on their ability to resist environmental stress conditions. From comparative omics data, we first identify the possible molecular mechanisms, and then we experimentally test functions and phenotypes. We do this through comprehensive data interpretation from omics analysis, ultrastructure, classical biochemical approaches, metabolic modeling, and genetic manipulation.
Reconstruction and Curation of Genome-scale Metabolic Models
The Srivastava Group is interested in applying quantitative systems biology to understand gene regulatory networks, metabolic networks, and signaling networks. Machine learning algorithms in conjunction with reaction kinetics theory are used extensively to mine experimental data and various publically available databases (e.g. GenBank, PubMed, etc.) to elucidate network architecture, discover new pathways/interactions, and generate novel hypotheses. Potential applications include more robust treatment of infectious diseases and cancer, as well as advancing and optimizing industrial biotechnology processes.
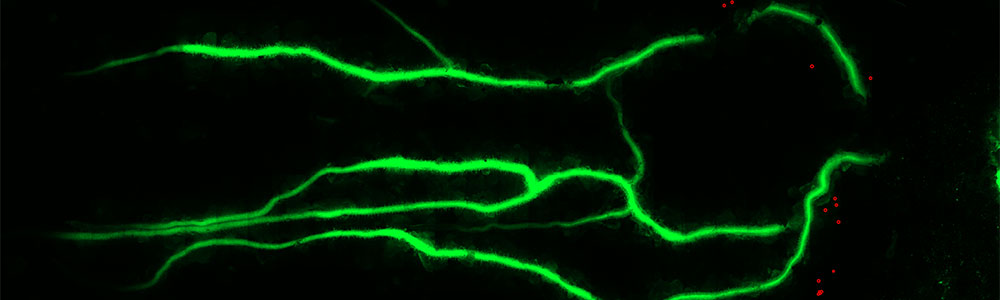
Understanding and monitoring host-microbe interactions in complex environments
The Gage lab uses a combination of traditional genetics, molecular biology, biochemistry microscopy, and genome-scale technologies to study the interactions of microbes with plants and various kinds of protists. In addition, we develop tools for monitoring complex environments at small spatial scales. Areas of particular interest are: 1) cell-cell communication, 2) signal transduction, 3) the assembly and function microbial communities that contain both prokaryotes and protists and 4) developing biosensor technologies.