Functional Genomics
Moving from genomes to functional information including proteomics and transcriptomics studies.
Biofuel generation through design and analysis of a genetically tractable extreme thermophile
Dr. Noll’s laboratory studies the physiology and molecular genetics of hyperthermophilic bacteria and archaea to shed light on their mechanisms of evolution, cellular structure, and potentials for biofuel generation. Strictly anaerobic, extremely thermophilic bacteria belonging to the group Thermotogales are the primary organisms under study. Our evolutionary studies focus on how the evolution of sugar transporters including their horizontal transfer among bacteria and archaea affects the binding properties of their proteins. Another research focus involves the unique cell envelope found in the Thermotogales, a structure with features perhaps reminiscent of cell barriers found in ancestors of bacteria and archaea. Finally a new effort to engineer thermophiles for use in biofuels generation is underway, an effort that seeks to address some of the major impediments to biomass utilization. Full genome sequences for several members of the Thermotogales have become available due to the efforts of our lab and this information is being used to guide these studies. Genomic and proteomic approaches are being used for these projects.
Determining biomolecular function of archaeal metabolism
Currently, Archaea diversity is analyzed principally at the (meta)-genomic level, providing valuable information on their metabolic and ecological potential; however, the function of many genes is still unknown. The Microbial Ecophysiology Lab (Santiago-Martinez Lab) is evaluating the gene expression and characteristics of some gene products (proteins). Our goal is to evaluate the function of biomolecules involved in energy metabolism and anabolic pathways of methane-producing archaea (also known as methanogenic archaea or methanogens).
Genomics, proteomics and transcriptomics of bacteria (actinomycetes) that infect higher plants and fix atmospheric nitrogen; diversity of fungi associated with surface ripened cheese
Benson's research interests include microbial biogeography, including the molecular and genomic interactions of bacteria with their environment, and the genomic evolution controlling distribution of microbes in several environments. The specific areas of interest include plant-microbe and insect-microbe microbiomes, and the distribution and diversity of microorganisms associated with cheese.
Microbial Analysis, Resources and Services (MARS) facility
The Microbial Analysis, Resources and Services (MARS) facility supports the research community of the University of Connecticut specializing in the analysis of microbial samples and high-throughput processing of nucleic acids. Examples include the characterization of microbiomes (Bacterial V4, Bacterial EMP, Fungal ITS, or custom amplicon), sequencing of small genomes, 96-well and 384-well PCR setup or DNA quantification and other automated liquid handling applications. Services are available a la carte, ranging from fee-for-service to unassisted use of the equipment by trained and certified users.
Mycoplasma gallisepticum genes and transcriptional pathways critical for interaction with, colonization of, and pathogenesis on host respiratory epithelium.
Steven Geary’s research interests are microbe-host interactions and evolution of microbial virulence, specifically involving bacterial pathogens of the genus Mycoplasma and primarily Mycoplasma gallisepticum; both in its natural poultry host and in the novel host, the American house finch. The lab uses comparative and functional genomics, transcriptomics, and in vivo models to investigate mechanisms of mycoplasmal pathogenesis, vaccine design, and virulence evolution. Website: http://patho.uconn.edu/people/faculty/indeste.html
Transcriptomic and metatranscriptomic studies using high-throughput sequencing
Senjie Lin's lab studies molecular ecology, evolution, and functional genomics of marine algae. Current research focuses on genome and transcriptome sequencing and analyses on dinoflagellates and other algae using high-throughput techniques.
Understanding and monitoring host-microbe interactions in complex environments
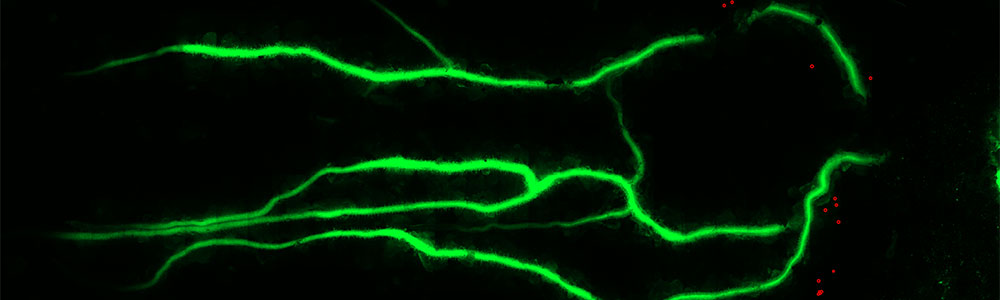
The Gage lab uses a combination of traditional genetics, molecular biology, biochemistry microscopy, and genome-scale technologies to study the interactions of microbes with plants and various kinds of protists. In addition, we develop tools for monitoring complex environments at small spatial scales. Areas of particular interest are: 1) cell-cell communication, 2) signal transduction, 3) the assembly and function microbial communities that contain both prokaryotes and protists and 4) developing biosensor technologies.
Understanding how beneficial symbioses are established and maintained
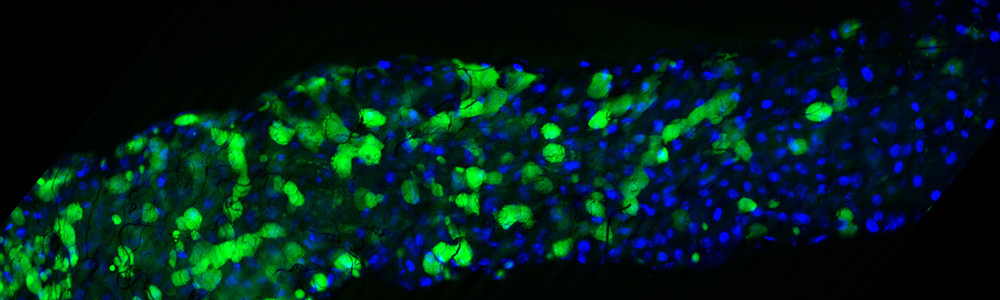
The Nyholm laboratory explores beneficial host/microbe interactions, ultimately trying to understand the mechanisms by which bacterial symbionts and their animal hosts communicate and how the host’s innate immune system might influence these interactions. We are currently studying how these complex associations are established, regulated and maintained. We use the model association between the Hawaiian bobtail squid, Euprymna scolopes, and the bioluminescent bacterium Vibrio fischeri, to to understand mechanisms of host/symbiont specificity. We are also exploring a bacterial consortium that is found in a reproductive gland of female squid and is hypothesized to provide antimicrobial and/or antifouling compounds in order to protect the developing, externally laid embryos.
Understanding how genetic variation is distributed within and between haloarchaeal species and how molecular mechanisms for gene transfer work in Archaea
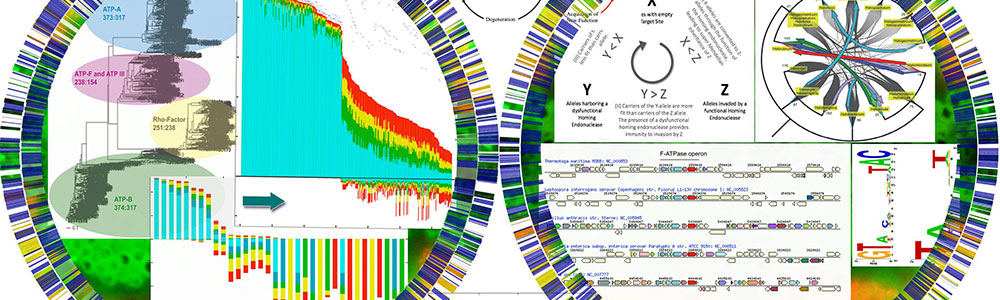
Papke lab Although we recognize names of microorganisms like Escherichia coli and Bacillus subtilis, our ability to classify strains into ³natural kinds² is rigorously tested by the observation that genetic variation is frequently shuttled across so-called species boundaries. Also, much of what we know about species comes from well-studied pathogenic bacteria, which are often classified by the disease that they cause (e.g., Bacillus anthracis or Neisseria gonorrhoeae). This is important to doctors and their patients but the Earth¹s biomass and diversity is comprised mainly of prokaryotes, the vast majority of which do not cause disease. Combined, these observations suggest that our understanding of species evolution is relatively shallow and that our current standards of classification are biased and difficult to apply to most microbes. The main goal of our research is to use evolution and ecology theory in combination with approaches like genomics, metagenomics and population genetics to investigate intra and inter species variation, gene flow and genetic relationships for non-pathogenic (e.g., environmental) prokaryotes. We concentrate on hypersaline adapted Archea (Haloarchaea) as model organisms because they live in island-like habits which aid in simplifying or sorting the evolutionary forces that effect their distributions, adaptations and variation.