Eukaryotic Microoganisms
Study of protists, fungi, algae and other microscopic organisms with nuclei.
Diversity and dynamics of planktonic ciliates - what can next-generation sequencing technologies tell us?
George McManus is interested in microbial eukaryotes and their roles in marine food webs. He is focused mostly on ciliates, which are among the most abundant herbivores in the sea. These organisms often practice a kind of mixotrophy in which chloroplasts from ingested food are retained and used to provide an energy subsidy for growth. His lab has successfully cultivated a number of these fastidious organisms. Using local isolates, he uses isotopes of C and N to study their metabolism. He is also interested in the biogeography of ciliates and uses DNA sequencing methods to evaluate their distributions and diversity.
Ecology and evolution of algae using molecular and phylogenetic approaches.
Senjie Lin's lab studies molecular ecology, evolution, and functional genomics of marine algae. Current research focuses on genome and transcriptome sequencing and analyses on dinoflagellates and other algae using high-throughput techniques.
Fungus-Growing Ants and their Microbial Symbionts
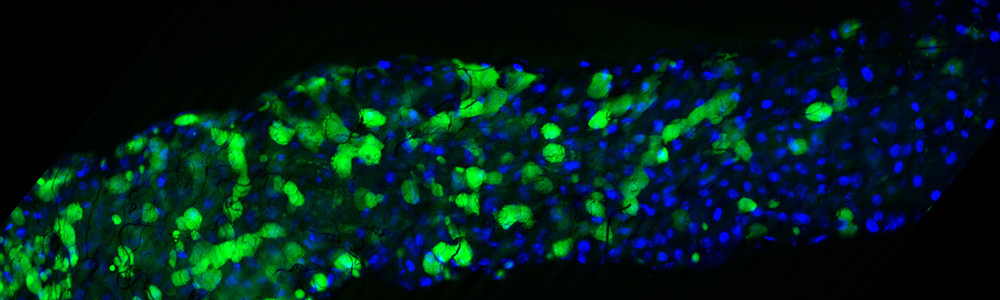
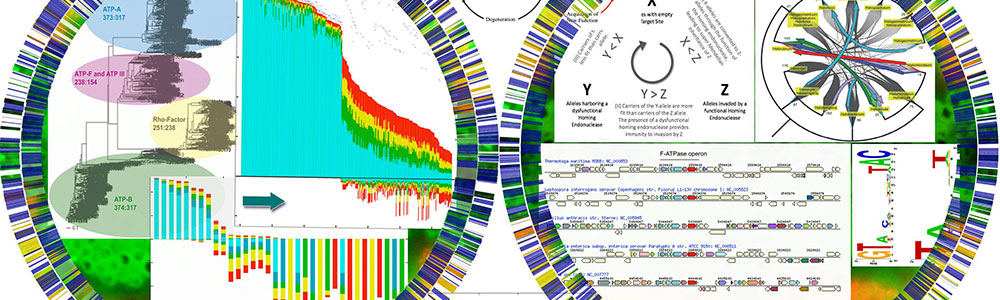
Jonathan Klassen studies microbial community ecology, especially using the fungus-growing ant symbiosis as a model system to understand how microbial interaction networks evolve. This symbiosis includes the invertebrate host and its multiple fungal and bacterial mutualists and pathogens, each with its own unique population structure and ecology. The Klassen lab particularly uses culture-dependent and -independent genomics coupled with comparative phenotyping and chemical biology to understand how the differing ecologies and population structures of each microbe governs the stability and function of the communities in which they reside.
Genomics, proteomics and transcriptomics of bacteria (actinomycetes) that infect higher plants and fix atmospheric nitrogen; diversity of fungi associated with surface ripened cheese
Benson's research interests include microbial biogeography, including the molecular and genomic interactions of bacteria with their environment, and the genomic evolution controlling distribution of microbes in several environments. The specific areas of interest include plant-microbe and insect-microbe microbiomes, and the distribution and diversity of microorganisms associated with cheese.
Microbial systems influenced by micro-scale habitat features
Leslie Shor’s research group studies microbial systems influenced by micro-scale habitat features. We have been developing passive protist samplers using microfluidic device technology. The motivation for the work is that biogeography of protozoa is poorly understood, yet protozoa play vital roles in global carbon cycling and mobilization of pollutants to higher trophic levels. Our microfluidic samplers link form, function, and taxonomy of protozoa communities in the field. Traps are designed to selectively trap protozoa based on cell size and plasticity, motility mode, and are deployed in a range of important natural environments.
Study of Trypanosoma brucei, which also serves as a model organism for other kinetoplastid parasites such as Trypanosoma cruzi and Leishmania spp.
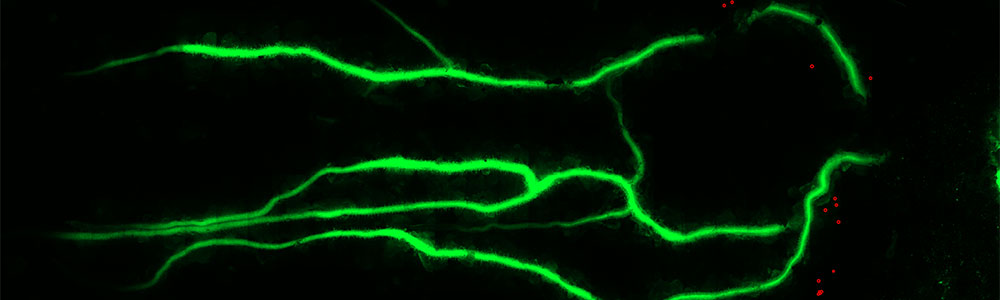
Arthur Günzl’s research team focuses on gene expression mechanisms in the eukaryotic, unicellular parasite Trypanosoma brucei which differ substantially from the correponding mammalian processes, depend on extremely divergent protein factors, and enable the parasite to evade the mammalian immune system by Antigenic Variation. We use biochemical, genetic and imaging techniques to study these processes and factors. A particular strenght of our group is the purification of protein complexes by tandem affinity purification.
Understanding and monitoring host-microbe interactions in complex environments
The Gage lab uses a combination of traditional genetics, molecular biology, biochemistry microscopy, and genome-scale technologies to study the interactions of microbes with plants and various kinds of protists. In addition, we develop tools for monitoring complex environments at small spatial scales. Areas of particular interest are: 1) cell-cell communication, 2) signal transduction, 3) the assembly and function microbial communities that contain both prokaryotes and protists and 4) developing biosensor technologies.