Genome Evolution
Investigating the evolution of genes, including the role of transfer of DNA, gene family expansion, and molecular parasites.
Development of computations tools to study gene family and genome evolution
Mukul Bansal’s research group develops new computational methods, efficient algorithms, and powerful software tools to help answer fundamental questions in molecular evolution. He is especially interested in problems related to understanding gene family and genome evolution through gene duplication, horizontal transfer, and loss, and in reconstructing highly accurate gene trees and species trees for both eukaryotes and prokaryotes.
Genomics, proteomics and transcriptomics of bacteria (actinomycetes) that infect higher plants and fix atmospheric nitrogen; diversity of fungi associated with surface ripened cheese
Benson's research interests include microbial biogeography, including the molecular and genomic interactions of bacteria with their environment, and the genomic evolution controlling distribution of microbes in several environments. The specific areas of interest include plant-microbe and insect-microbe microbiomes, and the distribution and diversity of microorganisms associated with cheese.
History of genome evolution (expansion) in dinoflagellates, using correlation between gene copy number and genome size
Senjie Lin's lab studies molecular ecology, evolution, and functional genomics of marine algae. Current research focuses on genome and transcriptome sequencing and analyses on dinoflagellates and other algae using high-throughput techniques.
Inferring and Testing the Genome Evolution of Microbial Symbionts
Jonathan Klassen studies microbial community ecology, especially using the fungus-growing ant symbiosis as a model system to understand how microbial interaction networks evolve. He uses genomic sequencing of both cultured isolates and environmental samples to understand the evolutionary history and distribution of these symbionts with high phylogenetic resolution. The Klassen lab also combines genomic analyses with comparative phenotyping and molecular biology to test inferred evolutionary reconstructions, with particular focus on evolution within multipartite communities.
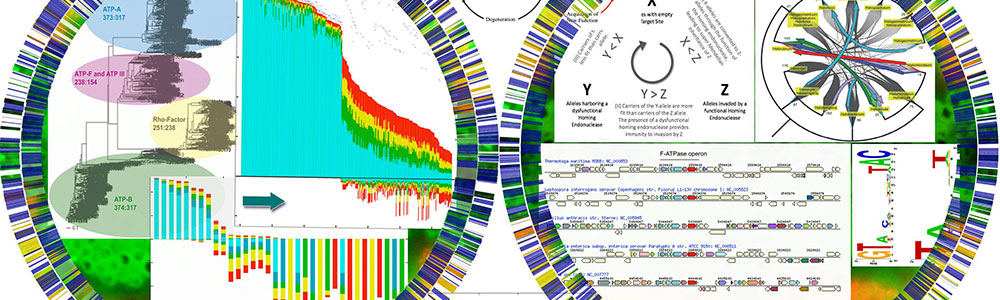
Reconstruction of ancestral proteins to examine the evolution of proteins inherited as a result of horizontal gene transfer
J. Peter Gogarten's lab studies the evolutionary histories of genes and organisms. Currently, the main research focus is comparative genomics and horizontal gene transfer. He also studies the early evolution of life (early expansion of the genetic code, gene family expansions, traces of extinction events in the molecular record) and the evolution of molecular parasites (inteins and homing endonucleases).
Reconstruction of the reticulated genome phylogeny from genomic data.
J. Peter Gogarten's lab studies the evolutionary histories of genes and organisms. Currently, the main research focus is comparative genomics and horizontal gene transfer. He also studies the early evolution of life (early expansion of the genetic code, gene family expansions, traces of extinction events in the molecular record) and the evolution of molecular parasites (inteins and homing endonucleases).
Reticulate evolution. The gene tree and species tree problem (focus on incomplete lineage sorting).
Yufeng Wu is interested in Computational Biology and Bioinformatics. Currently, the main research is mainly on computational problems in population genomics, phylogenetics and high-throughput sequencing.
Understanding how genetic variation is distributed within and between haloarchaeal species and how molecular mechanisms for gene transfer work in Archaea
Papke lab Although we recognize names of microorganisms like Escherichia coli and Bacillus subtilis, our ability to classify strains into "natural kinds" is rigorously tested by the observation that genetic variation is frequently shuttled across so-called species boundaries. Also, much of what we know about species comes from well-studied pathogenic bacteria, which are often classified by the disease that they cause (e.g., Bacillus anthracis or Neisseria gonorrhoeae). This is important to doctors and their patients but the Earth¹s biomass and diversity is comprised mainly of prokaryotes, the vast majority of which do not cause disease. Combined, these observations suggest that our understanding of species evolution is relatively shallow and that our current standards of classification are biased and difficult to apply to most microbes. The main goal of our research is to use evolution and ecology theory in combination with approaches like genomics, metagenomics and population genetics to investigate intra and inter species variation, gene flow and genetic relationships for non-pathogenic (e.g., environmental) prokaryotes. We concentrate on hypersaline adapted Archea (Haloarchaea) as model organisms because they live in island-like habits which aid in simplifying or sorting the evolutionary forces that effect their distributions, adaptations and variation.